Network
Collaborating across Europe to train the leaders of next-generation cancer research
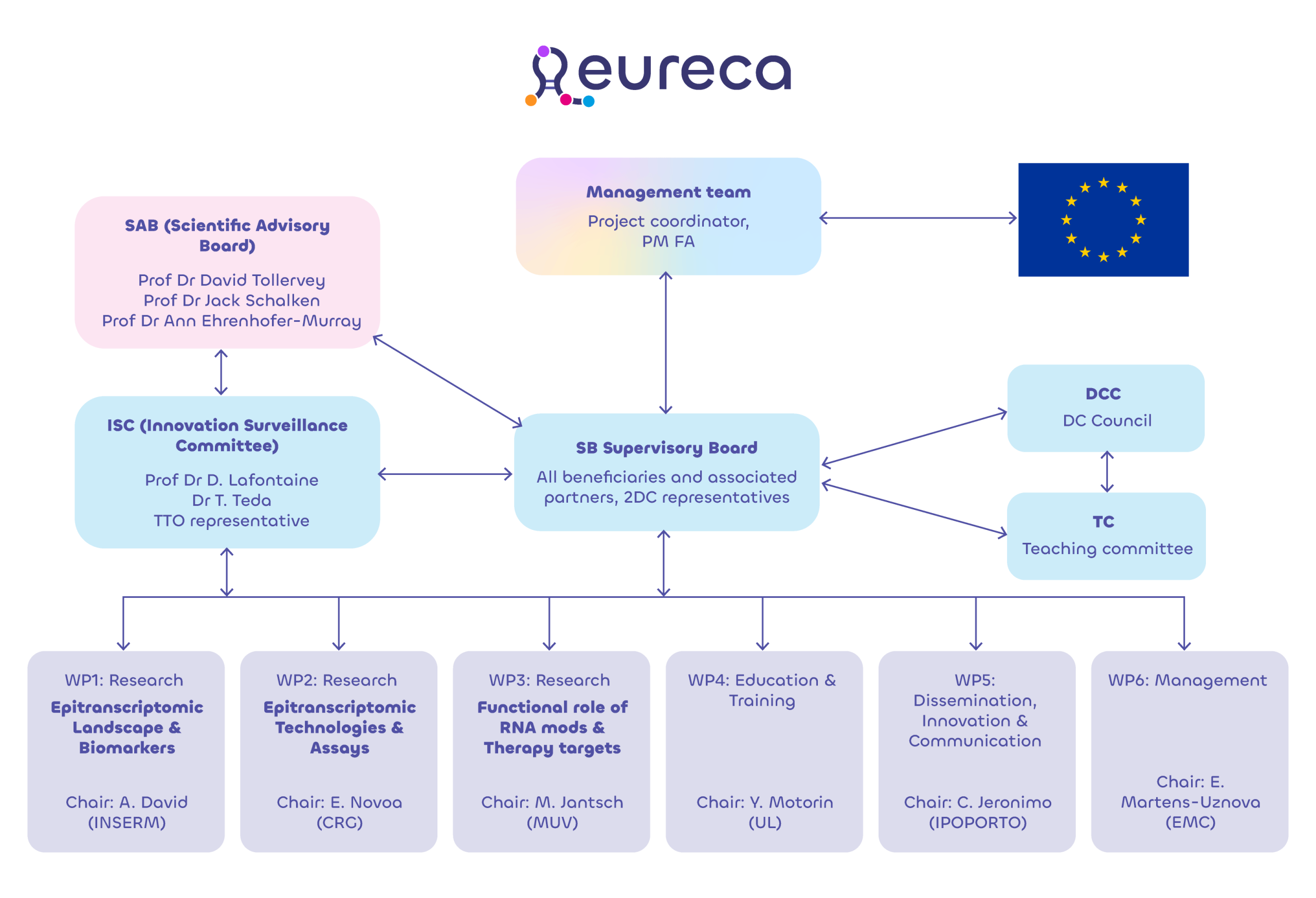
Team
Experts driving research, mentorship, and innovation
Principal Investigators
Doctoral candidates
Supervisory board
Principal Investigators

Prof Carmen Jeronimo
Cancer Biology & Epigenetics
EAU-ESUR and IPOPORTO
Doctoral candidates
Supervisory board


EURECA has received funding from the European Union’s (EU) Horizon Europe research and innovation programme under the Marie Skłodowska-Curie (MSCA) Grant agreement No 101226733.
Funded by the European Union. Views and opinions expressed are however those of the author(s) only and do not necessarily reflect those of the European Union or Horizon Europe MSCA-DN. Neither the European Union nor the granting authority can be held responsible for them.
© 2025 EURECA | Powered by Scienseed | Cookies Policy | Privacy Notice
Dr Elena Martens-Uzunova
As a molecular biologist my scientific credo says that the prerequisite to better detection and treatment of cancer is thorough understanding of the processes that occur in healthy cells. My research is focused on noncoding RNA and RNA modifications as modulators of RNA processing and function. I investigate how changes in RNA affect tumor growth and aggressiveness and how these changes can be used to develop better minimally-invasive liquid biopsy biomarkers and new therapeutics for prostate cancer.
Key words: liquid biopsy, minimally-invasive, extracellular vesicles, EV, exosomes, biomarker, RNA, snoRNA, tRNA, tRF, miRNA, urine, prostate cancer
FEATURED PROJECTS
Prof Michael Jantsch
The Jantsch lab studies RNAs and their maturation processes. Specifically, we care about the timing and interplay of transcription, splicing and RNA modifications and how these events affect the fate and function of RNAs.
A strong focus lies on the deamination of adenosines to inosines, which is an abundant modification in mRNAs. Inosines are being interpreted as guanosines by most cellular machineries and therefore affects the coding potential, splicing and immunogenicity of modified RNAs. We aim at understanding the patterns that affect immunogenicity of modified RNAs but also changes that occur to the proteome in response to adenosine deamination.
Key words: RNA maturation & processing, A to I modification, innate immunity
FEATURED PROJECTS
Alexandre David
Our goal is to advance the emerging fields of epitranscriptomics and translational control in cancer biology. We aim to redefine our understanding of gene expression regulation at the post-transcriptional level and translate these discoveries into novel clinical opportunities for personalized medicine. Each project begins with a clinical question and is dedicated to developing solutions for cancer detection, patient stratification, and treatment. As part of the S.M.A.R.T. consortium*, the TRANSLACORE European network and the GDR «RNA», we combine disciplines -such as chemistry, biology, bioinformatics and physics- and develop unique tools and techniques to produce innovative research at the crossroads of basic, applied and clinical sciences.
*SMART encompasses several teams from Montpellier: A. David (IRCM, epitranscriptomics), E. Rivals (LIRMM, bioinformatics), C. Hirtz (PPC-IRMB, analytical chemistry), M. Smietana (IBMM, oligo chemistry)

Prof Carmen Jeronimo
The Cancer Biology and Epigenetics Group of IPO Porto Research Center aims to advance patient-centred cancer research by integrating epigenetics, epitranscriptomics, tumour metabolism, and immunology to improve diagnosis and treatment.
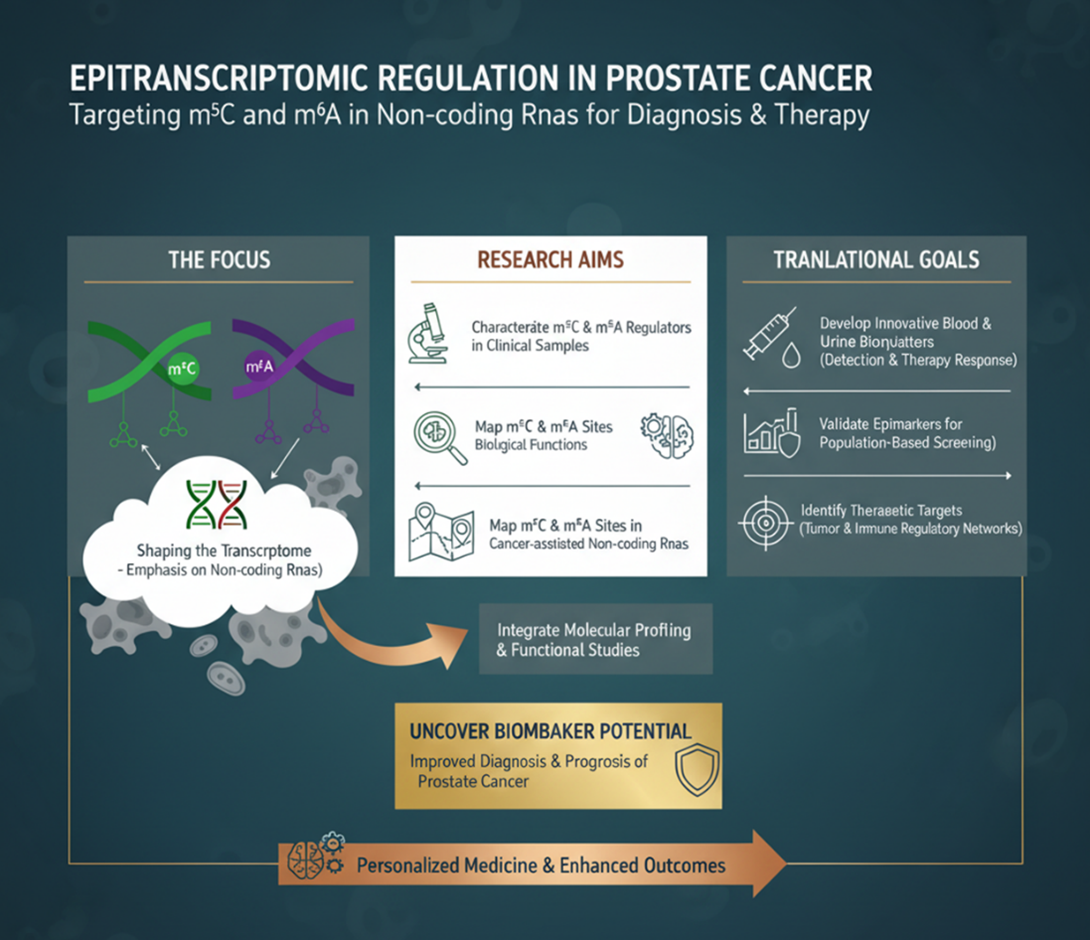
Regarding Epitranscriptomic Regulation, we focus on exploring the roles of m⁵C and m⁶A modifications in shaping the transcriptome of primary and metastatic prostate cancer, with particular emphasis on non-coding RNAs. Our work aims to characterize m⁵C and m⁶A regulators in clinical samples, elucidate their biological functions, and map m⁵C sites in cancer-associated non-coding RNAs. By integrating molecular profiling and functional studies, we seek to uncover the biomarker potential of m⁵C- and m⁶A-regulated non-coding RNAs for improved diagnosis and prognosis of prostate cancer.
We develop innovative blood and urine biomarkers for cancer detection and therapy response, validate epimarkers for population-based screening, and identify therapeutic targets within tumour and immune regulatory networks.
Through interdisciplinary and translational approaches, CBEG strives to enable personalized medicine and enhance clinical outcomes for patients with hard-to-treat cancers.
Prof Denis Lafontaine
The Université libre de Bruxelles (ULB) is a leading public research university located in Brussels, Belgium. Founded in 1834, it is internationally recognized for its commitment to academic excellence, critical thinking, and freedom of inquiry. With over 30,000 students and a vibrant international community, ULB offers a broad range of programs across all major disciplines and is home to numerous cutting-edge research institutes. The university fosters innovation, interdisciplinarity, and openness, making it a key player in European and global higher education. The Lafontaine lab is part of the Brussels South Charleroi BioPark, a ULB biotechnology and research campus located in Gosselies, near Charleroi.
The RNA Molecular Biology Laboratory at the Université libre de Bruxelles (ULB), Biopark campus, led by Denis Lafontaine, investigates the fundamental mechanisms of ribosome biogenesis, RNA processing and modification, and nucleolar organization. By combining molecular genetics, biochemistry, and high-resolution sequencing approaches, the lab explores how ribosome production is regulated in health and disease. A strong focus is placed on understanding the molecular basis of ribosomopathies such as Diamond-Blackfan anaemia and on developing therapeutic strategies to target nucleolar dysfunction.
Dr Eva Maria Novoa Pardo
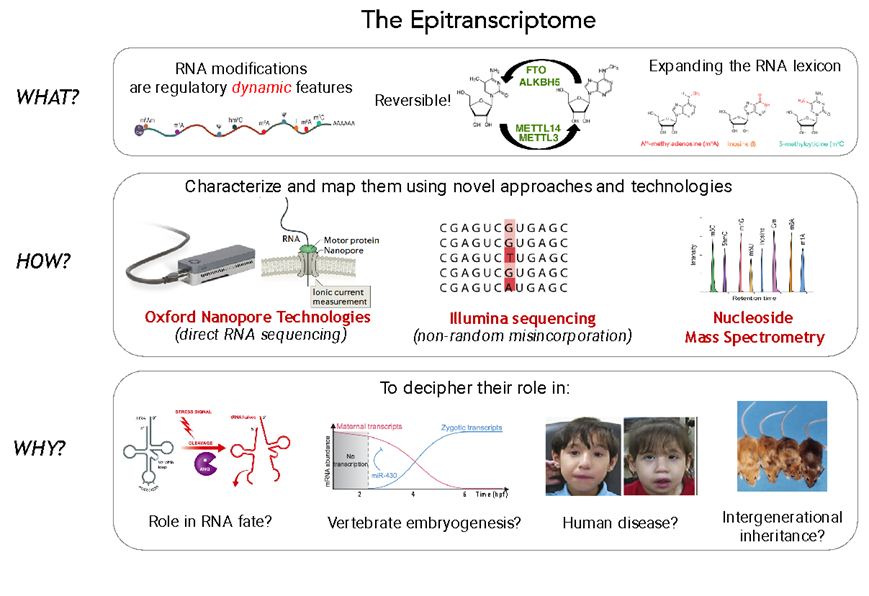
A current major challenge in biology is to understand how gene expression is regulated with surgical precision in a tissue-dependent, spatial and temporal dimension. Historically, genome-wide studies of gene expression have typically measured mRNA abundance rather than protein synthesis, in large part because such data are much easier to obtain. However, the correlation between mRNA levels and protein abundance is as low as r=0.35-0.40, suggesting that transcriptional regulation alone is not sufficient to unveil the complex orchestration of gene expression. In the last few decades, the scientific community has started to acknowledge the pivotal role that post-transcriptional regulatory mechanisms play in gene expression, however, we are still far from understanding how gene expression is finely tuned and regulated across tissues and conditions, suggesting that we are missing variables in the equation.
In our laboratory, we want to understand how gene expression is regulated with surgical precision, in a tissue-dependent, spatial and temporal dimension. To this end, we are employing a combination of experimental (RNASeq, polysome profiling, mouse/cell knockouts, Oxford Nanopore direct RNA sequencing) and computational techniques (NGS data analysis, algorithm development, machine learning), to unveil the secrets of three post-transcriptional regulatory layers: the epitranscriptome, RNA structure and ribosome specialization.
Our laboratory has pioneered the use and development of direct RNA nanopore sequencing technologies to study RNA modifications. Specifically, we have pioneered the development of novel algorithms to map and quantify RNA modifications (Liu et al., Nat Comm 2019, Begik et al., Nat Biotech 2021; Cruciani et al., Genome Biol 2025), we have optimized the library preparations to make the technology applicable to low-input samples (Pryszcz et al., Genome Res 2025) and we have expanded its applicability beyond coding mRNAs (Begik et al., Nature Methods 2022; Lucas et al., Nature Biotech 2024). We are now applying this technology to: i) decipher the role that RNA modifications play in sperm formation, maturation and intergenerational inheritance; ii) dissect how RNA modification dysregulation leads to human disease; and iii) bring this technology to the next level to apply it for rapid diagnosis of cancer, and classification of cancer types.
Prof Mark Helm
The Helm Group’s research focuses on RNA modifications of natural or synthetic origin. We integrate disciplines from chemistry, biology, physics, bioinformatics, and pharmacy to advance analytical and biosynthetic research on naturally occurring and therapeutic RNA.

Prof Alessandro Provenzani
The Laboratory of Genomic Screenings at the Department CIBIO focus on the development of novel assays, either phenotypic or biochemical, to be used in high-throughput screens using siRNA/CRISPR libraries or small molecules. The main focus is on the post-transcriptional area of gene regulation in diseases such as cancer and neurodegenerative ones. The aim is to identify target genes that are modulators of the disease, characterize their molecular mechanism of action, and develop an interventional approach. Our last efforts have been to screen for RNA-binding protein inhibitors, RAN translation inhibitors, and immunological modulators. After hit (gene or small molecule) identification, we make a strong effort to elucidate the mechanism of action of the target gene in the specific disease. We accomplish this goal by the synergistic utilization of classical biochemical/molecular biology techniques, genome-wide analyses, and high-throughput technologies. To name some of the projects, we have pursued the identification and development of disruptors of the RNA-binding proteins (HuR, YTHDF), activators of HSPB8, and modulators of RAN translation.
Prof Yuri Motorin
The Lab
The Redox Multi-scale Enzymology and Epitranscriptomics team focuses on fundamental mechanistic research and molecular engineering involving two classes of biologically relevant macromolecules (RNA and enzymes). The goal is to gain a better understanding of the fundamental physiological processes associated with these macromolecules while also developing their potential for valorization, particularly in human health.
The Core Facility
The Epitranscriptomics and Sequencing (EpiRNA-Seq) Core Facility is built on the expertise of the UMR7365 IMoPA in the field of RNA molecular biology to promote its main collaborative services.
Located on the Brabois-Santé campus of the University of Lorraine in Nancy, it provides to the academic and industrial scientific community high-tech resources (Illumina next generation sequencers). It is part of the European COST EPITRAN network and regularly organises training activities.
The core facility has recognized technical and methodological know-how to contribute to various projects in Epitranscriptomics (RiboMethSeq, AlkAniline-Seq, HydraPsiSeq) to address various scientific questions such as dynamics of post-transcriptional modifications in RNAs.
The EpiRNA-Seq Core Facility operates mainly in collaborative mode (involvement of members of the core facility in the steps of your sequencing project from the conception of the experimental design to the statistical analysis of the data) but the delivery mode (mainly sequencing of libraries prepared by you without involvement of the platform) is also possible under certain conditions. The EpiRNA-Seq Core Facility has a differential pricing system (Université de Lorraine members, academics and industrialists).
Dr Julien Lagarde
Flomics Biotech, S.L. is a start-up company based in Barcelona, operating in the transcriptomics and liquid biopsy field. Flomics aims to develop a multi-cancer early detection (MCED) test from a standard blood sample. At the core of Flomics’ business model is our proprietary technology focused on the analysis of cell-free RNA (cfRNA), a highly informative biomarker for understanding various disease states. Our platform combines next-generation sequencing (NGS) technology with machine learning to study cfRNA profiles in plasma. The company has developed a proprietary in vitro pipeline for cfRNA analysis from liquid biopsies, including blood collection, RNA isolation, DNase treatment and library preparation.
Flomics has also built a custom in silico pipeline to process the cfRNA-Seq data. This pipeline conducts quality control, trimming, read alignment, and gene quantification using the latest human genome annotations. It generates a wide range of features per sample, including gene abundance and RNA splicing patterns, with over 100.000 potential features or biomarkers identified.
Flomics collaborates closely with hospitals, biobanks, and other international partners to ensure the successful implementation of its innovation across Europe, ultimately improving patient outcomes.
Prof Ernesto Picardi
The Bioinformatics and Omic Sciences lab, led by Prof. Ernesto Picardi, has extensive expertise in molecular biology, bioinformatics, and Omic Sciences. It participates in various projects focused on developing bioinformatics tools for analysing large sequencing datasets, particularly methods to interpret the epitranscriptome. Its widely used tools, such as REDItools and REDIportal, serve as primary resources globally for profiling A-to-I RNA editing in high-throughput sequencing data. Through the national ELIXIR node, the lab has access to advanced omics platforms- including Illumina, PacBio, and Oxford Nanopore- and the RECAS Datacenter, one of Italy’s most powerful bioinformatics computing centers, to support omics data production and analysis.
Prof Eric Rivals
Our lab
Our lab conducts research in computer science and bioinformatics, more precisely on algorithms for biological data processing, on analysis of sequencing data, on inference and visualisation of evolutionary relationships. We develop and maintain software that implement most of the algorithms designed in our team and one of the first public, bioinformatic platform, named ATGC, which offers computational services and tools for life sciences. Our group is involved in collaborative multidisciplinary research projects in biology and life sciences. We actively collaborate with molecular and evolutionary biologists, physicians, oncologists, chemists by leveraging state-of-the-art computational approaches to investigate key biological and clinical questions.
In collaboration with the groups of Dr. Alex David at the Cancer Res. Institute of Montpellier (IRCM) and of Prof. Christophe Hirtz at Montpellier Univ Hospital (CHU), we have developed an original approach to identify biomarkers for cancer diagnostics, stratification, and disease follow-up, which has led to patents and to the creation of the start-up Cyberna.
About LIRMM / CNRS
The Laboratoire d’Informatique, de Robotique et de Microélectronique de Montpellier – LIRMM (Laboratory of Computer Science, Robotics and Microelectronics of Montpellier) is a major multi-disciplinary French research center located in the South of France. The LIRMM is a joint unit of Montpellier University and CNRS (and is thus affiliated to both). It conducts research in Computer Science, Microelectronics and Robotics and is organized along 3 departments comprising 19 international research teams.
The National Centre for Scientific Research (CNRS) is a major player in basic research worldwide, and the only French organisation active in all scientific fields. Its unique position as a multi-specialist enables it to bring together all of the scientific disciplines in order to shed light on and understand the challenges of today’s world.
Montpellier University
The University of Montpellier (French: Université de Montpellier) is a public research university located in Montpellier, in south-east of France. Since the establishement of its medical faculty in 1220, the University of Montpellier is one of the oldest universities in Europe and in the world. Over 50000 students currently follow a cursus among its 17 faculties, schools and institutes. The University is organised following its five major research domains: 1. Agriculture, Environment, Biodiversity, 2. Biology-Health, 3. Chemistry, 4. Social Sciences and 5. Mathematics, Informatics, Physics, Systems (MIPS). The nine doctoral schools are training circa 2200 PhD students counted over all disciplines.
Dr Virginie Marcel
The “Ribosome, Translation and Cancer” team led by Jean-Jacques Diaz is composed of 18 permanent members. The team has challenged the role of ribosomes in cancer, studying both quantitative alterations (i.e., hyper-activation of ribosome biogenesis) and qualitative alterations of ribosomes (i.e., alteration in ribosome composition). The main objectives of the RTC team were to study the role of ribosomes and translation in cancer by focusing on:
In this context, V Marcel’s group (EURECA partners: V Marcel, C Moyret-Lalle, S Durand) has developed a basic to translational research program that contributes to the establishment of the concept of ‘ribosome heterogeneity’ in the context of cancer by focusing on the main chemical modifications of rRNAs, the 2’O-ribose methylation (2’Ome). Our current objectives are to develop an integrative approach to decipher the combined role of all rRNA chemical modifications in translational regulations and in cancer progression, and thus to:

Dr Karin Garber
Dr. Karin Garber is a director at Open Science – Life Sciences in Dialogue, a non-profit association in Vienna dedicated to fostering dialogue between science and society. Since 2004, she has been leading the implementation and operation of the Vienna Open Lab, Austria’s first and largest public hands-on laboratory, which welcomes over 14,000 visitors each year.
At the Vienna Open Lab, Karin Garber combines her expertise in genetics, education, and science communication to create interactive learning experiences that make molecular biology and genetics accessible to people of all ages. She is involved in designing workshops, educational programs, and outreach activities that promote scientific literacy and public engagement with research.
In addition to her work at Open Science, Karin Garber lectures on Science Communication and Project Management at the University of Vienna. She has extensive experience in developing science education content, conducting teacher trainings, and mentoring early-career researchers in effective communication.
Within the EURECA project, the Open Science team supports the consortium’s PhD students in enhancing their communication and public engagement skills through interactive and practice-oriented training formats. These activities aim to equip early-career researchers with effective communication strategies and participatory approaches to connect their scientific work with society.